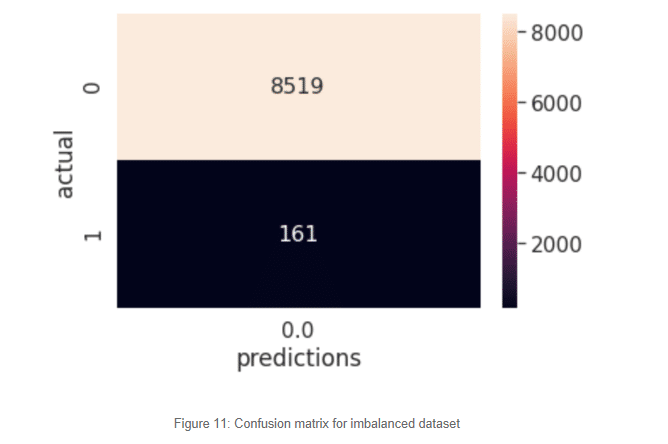
Something quite interesting is happening here. The class distribution is so skewed that our model has learned that the best way to classify our target variable is to predict everything as a negative case. Of course, it has seen over 30 thousand negative cases and just over eight hundred positive cases. At this point it’s almost like a random guess, the algorithm determines the best way to classify the never-seen-before examples is to mark them all as zeroes. This is exactly the problem we face with imbalance datasets. Those 161 cases predicted as healthy are most likely to develop a stroke and our model was not able to predict it. The cost of misclassification at this scale is impossible to intolerable.
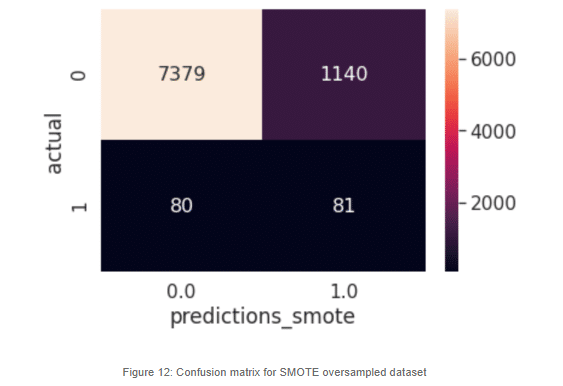
Now this looks a lot different than the earlier model. When increasing the minority class artificially, the model can make a better generalization. We can now predict almost half of the positive cases, which is an improvement. Still, there are a lot of misclassifications in the negative cases. There are a lot of negative cases that are being marked as possible stroke victims, but for our most sensitive cases, which are the actual positives, we can see that our classifier is doing a much better job.
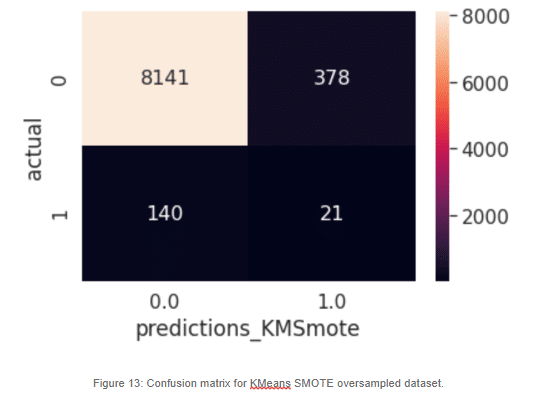
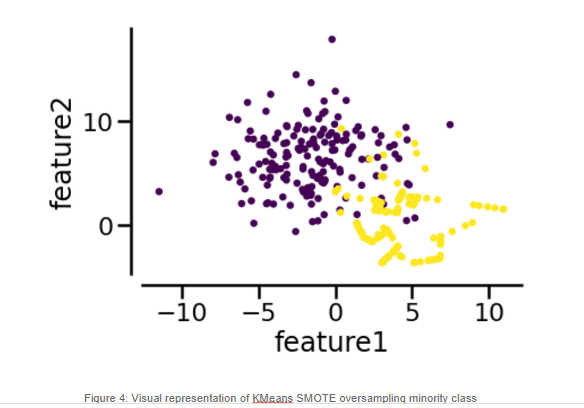
For our KMeans SMOTE variant, we can see that it performs worse than regular SMOTE. We can see a degradation of the false negatives. In this case, without further investigation, we cannot make a solid statement on why KMeans SMOTE is the lowest performer, but it may have to do with the fact that KMeans itself has its own critical Hyper-Parameter, k, which should be optimized. If a sub-optimal number of clusters is generated, perhaps our artificial samples won’t help our model. Supplemental tuning of the KMeans SMOTE and the model itself may help improve the performance of the classifier, however we will not address this in this article.
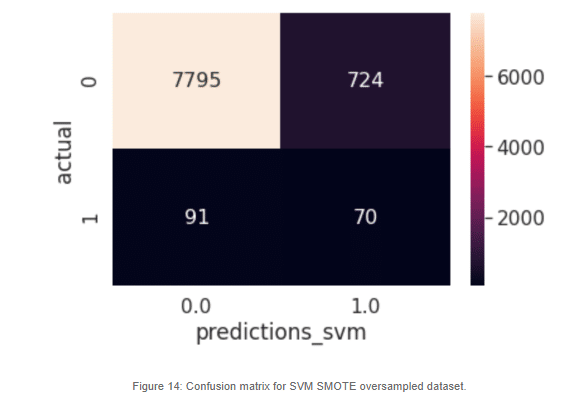
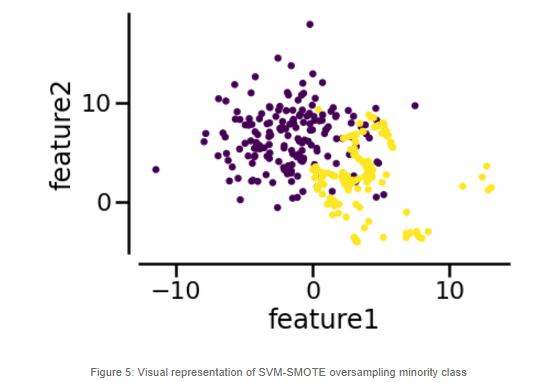
SVM-SMOTE resampled dataset produces a better result than the KMeans SMOTE variant when it comes to the positive cases. This could mean that the area where misclassification is happening is at the border of the two classes. We could further improve this model by tweaking the class distribution under sampling the majority class.
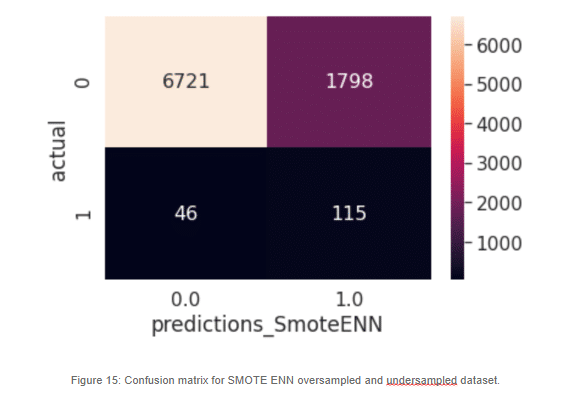
Lastly, we trained a classification model using both SMOTE oversampling for increasing the minority class and decreasing the majority class. Of the models described, this one produces the better results regarding the false negatives. Still there is a lot of misclassifications in the actual negatives, but for our most important cases, this model achieves better performance than its peers.
For the following section, we will discuss the different methods available for the different oversampling techniques discussed in previous chapters. We will focus on Imbalanced-Learn, an open source library relying on scikit-learn which provides tools to deal with classification with imbalanced datasets
Now that we have an imbalanced distribution, it’s time for splitting our data.
A very important note here: No matter which technique you use to overcome an Imbalanced Dataset, you must always apply it after partitioning your data in train-test.
If you oversample, do it on the training dataset, never on the validation dataset. Why? Well, because if we oversample on the validation dataset we will “contaminate” our model evaluation phase with data that will not reflect our reality. Remember, our objective is to build models that generalize over the observations and in the real world, for this kind of problems, it is normal that the minority class is underrepresented and appears in few cases and what we really need is to correctly predict these minority observations.
Having said this, let’s take a look at the way we partition our dataset:
# Split into train-test
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_resampled, y_resampled, test_size = .3, random_state=42)
Train_Test_Split is a great way to split our datasets. Is a method of Scikit-Learn and its main advantage is that it will not only split our dataset, but it will do a random shuffle too, that will ensure that our dataset is properly randomized and then split. Not shuffling the data before splitting may cause an abnormal proportion between classes in training and validation datasets, so it’s nice to have randomization and splitting together. In our example we define a validation size of 0.3, which means 30% validation and 70% training. This is usually the best practice.
Now that we have our imbalance dataset split into training and testing, it’s time to make use of our resamplers. We will demonstrate default SMOTE, KMeans SMOTE and SVM SMOTE.
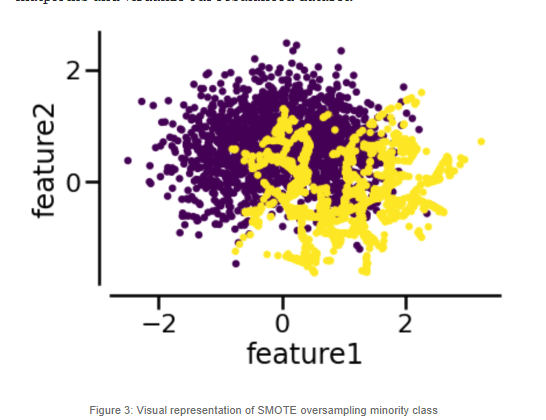
Remember that in its plain form, SMOTE will randomly generate samples in the segments connecting the K Nearest adjacent minority class instances and will repeat this process until the dataset is no longer imbalanced.
But sometimes we want to place synthetic samples with a different approach. We will now show you how to implement KMeans and SVM Smote, which relies on an unsupervised algorithm to determine where to place the synthetic samples in a more efficient way.
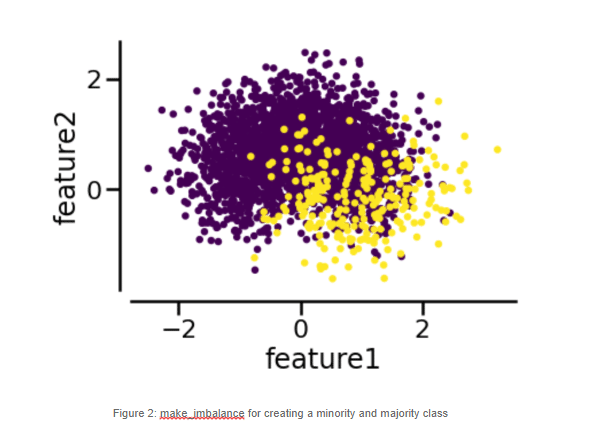
To properly demonstrate the following methods, we proceed to create a new artificial dataset, specially created for binary classification, using the make_blobs method. According to the official documentation, make_blobs is a method for generating isotropic Gaussian blobs, which are perfect for clustering purposes, and this is how we applied it:
# Use make_blobs to create dataset
from sklearn.datasets import make_blobs
X, y = make_blobs (n_samples = 500, centers = 2, n_features = 2, cluster_std = 3, center_box = (-7.0,7.0) , random_state = 42)# Convert to pandas dataframe for better handling
import pandas as pd
X = pd.DataFrame (data = X, columns= [“feature1”, “feature2”])
y = pd.DataFrame (data = y)# Use make_imbalance to create a skewed dataset
from imblearn.datasets import make_imbalance
X_resampled, y_resampled = make_imbalance (X, y, sampling_strategy = {0:250, 1:50}, multiplier = 0.1, minority_class = 1)# Split in train-test
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_resampled, y_resampled, test_size = .3, random_state=42)
Now, we are ready to apply our resampling techniques: SVM SMOTE and K-Means SMOTE.
from imblearn.over_sampling import KMeansSMOTE
sm = KMeansSMOTE(random_state=42)
X_over, y_overs = sm.fit_resample(X_train, y_train)
Previous snippet shows the use of KMeans SMOTE for synthetic sample generation. As you can see it follows the same structure. As result we get the following output: